KEYWORDS
Microbiota
Metabolomics
Genomics
Transcriptomics
Molecular technology
Irritable bowel syndrome
Abstract
Nutraceuticals and probiotics have been in focus in nutritional research because of their benefits to human health. These benefits, combined with the need for improvements, have resulted in a pursuit of new options to elucidate the mechanisms of action of bioactive compounds. Development of omics techniques have been able to fill in the knowledge gaps regarding the role of nutraceuticals and probiotics in human health. The integrated approach from different omics techniques is required to obtain the flow of molecular information from one ‘omics’ level to the other. The different omics techniques, genomics, transcriptomics, proteomics, and metabolomics used for studying nutraceuticals and probiotics and their influence on the host are discussed in this article.
Introduction
High throughput platform development and powerful bioinformatics’ tools in combination have contributed to our comprehension of the role of nutraceuticals, probiotics and other bioactive ingredients in human health. They have helped us study the biological and the cellular responses induced by these supplements, allowing us to validate and understand their health promoting capabilities. Current trend in the omics approach is using multiple omics platforms in collaboration, to achieve a comprehensive understanding of the nutraceuticals and probiotics functionality in human health. Different omics platforms help in deepening our knowledge of interaction of nutraceuticals, bioactives and probiotics, with the host and studying their mechanism of actions, safety concerns, efficacy and characterization including development of new health ingredients from traditional medicines (1). Plant-derived bioactive molecules, phytonutrients, consisting of flavonoids and phenolic compounds have an important function in human diet and nutrition. Genomics, transcriptomics, proteomics and metabolomics are main omics analyses that can be used for functional foods and nutraceuticals (Figure 1). Natural phytochemicals may affect signaling pathways and genome expression which can be studied using genomics (nutrigenomics) and transcriptomics. Functional foods and nutraceuticals like carotenoids, polyamines, folates, phenolics, flavonoids and non-flavonoid condensed tannins have been shown to modulate various cellular processes (2,3).
The human and animal gastrointestinal tract is inhabited by countless microorganisms. The total number of genes of these microbes are estimated to be almost 150 times the number of human genes. These microbial genes have key roles in physiological functions and health of the host. Genome mining has allowed us to determine properties such as acid tolerance, antimicrobial peptides production in possible probiotic strains, metagenomics moreover contributes to understanding the probiotic-microbiota interaction. Transcriptomic, proteomic, and metabolomic platforms have supported the profiling of host–microbe crosstalk and have contextualized the holistic effects induced by the supplementation.
Nutrigenomics analyzes the interaction of nutrients with human genomic sequences and studies its influence on host genomic expression and transcription (4,5). Multiple omics platforms are now a necessity to gain understanding of the mechanism of action, as well as the impact on metabolic pathways and influence on homeostasis by nutraceuticals and probiotics in humans. Nutrigenomics is built on understanding the functional sensitivity of genes to nutrients which is evident from the metabolic complications that may be caused in infancy to later years, due to nutritional deficiencies in mothers. For the purpose of this article the word nutraceuticals will be used to cover all dietary ingredients and supplements used for promoting human health.
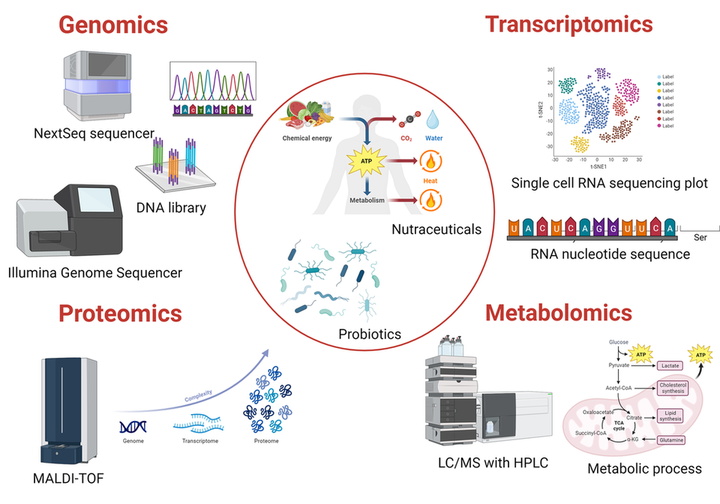
Figure 1. Integrated use of “omics” techniques can help achieve a broader understanding of the role of nutraceuticals and probiotics in human health.
Created with BioRender.com
Genomics
Genomics may involve studying structural genomics and functional genomics. Structural genomics determines the 3-D structures of proteins encoded by a given genome and includes a high-throughput method for structure determination through a combination of experimental and modeling approaches. A major part of genomics is sequencing the genomes of various organisms studying their genetic expression under various conditions. Functional genomics on the other hand attempts to describe gene and protein functions and their interactions.
In microbiology, genomics has allowed us to sequence diverse strains of multiple species, elucidating the pangenome for protein coding. Metagenomics on the other hand has allowed analyses of microbes, in particular the non-cultivable bacteria at stain levels (6). Genomic analyses have been used to determine the mechanisms of host-microbe interactions, including effects on cell signaling pathways, bile secretion, proteasome, cAMP signaling, thyroid hormone synthesis, and dopaminergic synapse pathways. For example, a recent study in 2020 used a genome-based approach to show presence of genes for lactic acid production, stress tolerance and antimicrobial peptides (7).
The DNA sequencing technology started from DNA microarrays to first generation Sanger sequencing, then came the second generation with massive parallel sequencing capabilities more commonly known as next generation sequencing with platforms such as 454 Genome sequencers (Roche Life Science, USA) and Illumina Genome Analyzer (Illumina, USA), and then the current third generation with long reads sequencing which has allowed sequencing whole genome so deep that mutational changes can be characterized enabled by Pacific Biosciences and Oxford Nanopore technologies (8).
Metagenomics and metatranscriptomics in conjunction have contributed to the development of an improved, new generation of probiotics. With these techniques we have a better understanding of the microbiome, and they can be used to generate valid hypotheses to determine the mechanism of dysbiosis progression in human body and its consequences (9). A large-scale genome wide analyses among lactic acid bacteria illustrated their diversity, prevalence and relationship to each other, and identified the link between gut and food microbiome (10). Metagenomics has been widely applied in the research of obesity, providing evidence for the important role of intestinal microbiota for obesity. There was a difference in relative abundance when the distal gut microbiota was compared for obese versus lean individuals, with increase in relative abundance of Bacteroidetes was observed when obese individuals lose weight (11). Metagenomics studies have also demonstrated an imbalanced microbiota composition in various diseases, for example Crohn's disease, necrotizing enterocolitis, polyposis or colorectal cancer, and type 2 diabetes (11).

Transcriptomics
The concept of transcriptomics is based on the central dogma theory where the genetic information flows from DNA to DNA (replication), from DNA to RNA (called transcription), and from RNA to protein (called translation). In this sequence the transcription results in the formation of a complete set of RNA consisting of ribosomal RNA, tRNA, mRNA and other non-coding RNAs. However, only mRNA carries the identities of individual genes and acts as bridges between genotypic information to the phenotypic expression of an organism. Transcriptomics includes the whole transcripts expressed from the genome, thus making it an ideal platform for studying the expression of various genes (12). Genetic expression measurements of genes in different tissues under various conditions at multiple time points provides information about gene regulation and reveals details of biologicals functions in an organism. It also provides information on previously unknown genes, hence transcriptomics analysis in its entirety has enabled us to understand trends in disease progression or health benefits downstream of a bioactive compound. Bioactive components from foods and dietary supplements can change gene expression by affecting the cellular pathways and metabolism, cell proliferation and differentiation and finally cell death and any imbalance in these processes can lead to development of diseases such as diabetes or cancer. Thus, the influence of nutraceuticals and other bioactive components on genetic expression at genome level is an important part of nutrigenomics investigations. Transcriptomic analysis has been employed in clinical trials to get an understanding of the effects of dietary interventions such as investigations for nutritional deficiency, intermittent fasting, or certain food components. Some of the most common pathways studied using transcriptomics in health promoting dietary interventions are glucose and insulin signaling, inflammation, oxidative phosphorylation, immune responses, and circadian rhythm (13). Transcriptomics was applied to study the internal response of cells to physical disturbances. It can also be applied to study the host-microbe interaction in understanding the effect of probiotics on host immune modulation for e.g., the effect of Bifidobacterium breve UCC2003 strain on strengthening the intestinal barrier by having an effect on the intestinal epithelial cells was demonstrated using transcriptomics. The strain influenced the epithelial cells by regulating gene expression and influencing metabolic pathways (14).
Before advancement in the next generation sequencing technologies, RNA microarrays were used. Some of the nutraceuticals and bioactives studied using microarray techniques for transcriptomics have been green tea catechins, vitamins, polyunsaturated fat, isoflavones, quercetins, anthocyanins, and arginine (1). Once RNA sequencing became available it was used to detect the abundance of RNA transcript in high throughput manner. Additionally, several other RNA sequencing platforms have also developed that involve tag based methods i.e. using one fragment to represent a transcript such as digital gene expression sequencing and 3’ end sequencing. Currently the transcriptomics technology is chip based which includes cDNA microarray and oligonucleotide chips, serial analysis of gene expression, and massively parallel signature sequencing and there are several other variations in the techniques which are beyond the scope of this article (8).
Similar to metagenomics, metatranscriptomics has been employed to determine the microbial composition and diversity from many ecological niches for e.g. researchers employed metatranscriptomics to study the complex interaction of the oral microbiome in biofilm assembly (15). Metatranscriptome analysis normally sequences the entire transcriptome from the microbial community and unravels the complex interactions between probiotics, the microbiome, and the host. It can give a clear picture of specific targets and precision application of probiotics, especially when coupled with other omics techniques such as proteomics and metabolomics (16).


Proteomics
Proteomics involves high-throughput analysis of proteomes in cells, tissues, or biological fluids which are being expressed by the genome at a specific growth phase and specific growth conditions including studying the post translational modifications (methylations, phosphorylation, or glycosylation). It allows us to elucidate and quantify the proteins associated with a cellular process such catalysis and stress response. The proteome is dynamic and varies depending on the cell type, its functional state and because of the alternative splicing and post translational modifications the ratio of genome to proteins expressed is much different (17).
Proteins involved in probiotic and gut microbiota functionality have been studied using proteomics (18). Proteomics and metabolomics in combination have been used to study the functions of probiotics at different stages of production process to determine the effect of production stressed on the potential of the probiotic strain (19). Proteomics is an attractive target for studying aging biomarkers as they determine the phenotype in host (20). Proteomics in nutritional research has provided knowledge about the interaction of the nutraceutical and protein regulation in host, effects on the expression of the proteins in response to the nutrient administration, and the possibility for identifying the biomarkers that are sensitive to the intervention being studied (21). Compared to genomics and transcriptomics there are much fewer studies on nutritional proteomic analysis. Some examples are identification of cellular protein targets in diet by protein profiles generated using cell culture studies using epithelial intestinal cell lines to study the effect of dietary ingredient, such as butyrate or flavonoid (1).
Proteomics techniques have progressed from gel-based techniques such as 2-DE (two-dimensional) gel electrophoresis to technologies such as mass spectrometry by evaporation of peptides and proteins by MALDI (matrix-assisted laser desorption/ionization) and ESI (electrospray ionization), multiple reaction monitoring, and multiplexed immunoassays. The techniques used in proteomics include protein microarray used to study cholesterol mediated change in protein expression, the 2D gel and MALDI-TOF-MS proteomic analysis techniques which were utilized to identify the prospective allergens from transgenic soybean and non-transgenic soybean (22).
Metaproteomics measures total expressed proteins and is considered to provide more precise functional information than genomics and transcriptomics-based predictions. MS based proteomics has been used to measure protein content from both host and microbiota (23), and has proven to be a powerful tool to determine host microbe interaction in a complex ecosystem. Meta proteomics has allowed an understanding of the community phenotypes (24) and since cells are mainly composed of proteins metaproteomics can help us understand the influence of community members to the community biomass (25).

Metabolomics
Metabolites are generally low molecular weight, less than 1500 Da, biological effector molecules with several functions such as their role in cellular metabolism, energy harvesting or they can be signaling molecules in an organism, maintaining cellular homeostasis. Metabolome is a collection of all metabolites in a cell other than the genome, transcriptome and proteome. There are two approaches to analyzing the metabolome; the targeted approach which involves searching and quantifying specific metabolites in known pathway to prove a specific biological hypothesis. Non-targeted metabolomics approach aims to identify de novo targets by quantifying as many metabolites as possible in a sample. Metabolomics is capable of detecting hundreds of metabolites in a biological system simultaneously, which reveals the health status of an organism pointing to its phenotype. Low molecular weight compounds such as peptides, carbohydrates, amino acids, nucleic acid metabolites, vitamins, organic acids, and minerals produced by microbial metabolism can be identified using metabolomic techniques.
Several methods are used to determine the metabolome including Fourier transform-infrared spectroscopy, Raman spectroscopy, well-known technique NMR spectroscopy and mass spectroscopy (MS)-based approaches such as liquid chromatography-MS, gas chromatography-MS among others. MS is quite feasible and popular for studying metabolome because of its ability to detect a wide spectrum of metabolites (1). The output from metabolomics techniques consists of measurements performed on subject under different conditions. These measurements can consist of digitized spectra, or concentrations of a certain metabolite. Pattern recognition methods and statistical programs such as principal components analysis and partial least squares can be employed to perform downstream analysis to extract meaningful information from the metabolomics output. Change in metabolites may occur due to disturbances in the living cells, irrespective of the physiological factors, thus the metabolome represents physiological or pathological status of organisms (8).
In nutraceutical research metabolomics can be used to provide information about biochemical deviations in response to dietary interventions. A study used 16S rRNA sequencing in combination with NMR analysis to show that metabolomics can used to correlated certain biological functions with taxonomy (26). Additionally, some studies show that secondary metabolites produced by probiotics influence the interaction of bacteria in community and causes attenuation of virulent pathogenic biomarkers (27). Metabolomics has also been used to show the changes that occur during the fermentation of foods and dairy products in the presence of lactic acid bacteria (16).
Conclusion and future directions
A single ‘omics’ technique is insufficient to fully understand biological processes clearly. However, the omics in combination are powerful techniques that can be used to study the effect and mechanism of nutraceuticals and probiotics on human health. As the techniques within each omics are advancing, our understanding of biological systems and the interaction and influence of dietary supplements to it is increasing. Care must be taken, though, to not over interpret output from these techniques. Instead, a systematic approach is needed and carefully designed hypotheses should be tested using omics as tools to get a clearer understanding. The omics-based approaches will guide the growth and development of nutraceuticals and probiotic selection in food and nutrition sciences.

References and notes
- Pandita D, Pandita A. Omics Technology for the Promotion of Nutraceuticals and Functional Foods. Front Physiol. 2022;13:817247.
- Islam SU, Ahmed MB, Ahsan H, Lee YS. Recent Molecular Mechanisms and Beneficial Effects of Phytochemicals and Plant-Based Whole Foods in Reducing LDL-C and Preventing Cardiovascular Disease. Antioxidants (Basel). 2021;10(5):784.
- Nayak SN, et al. Omics Technologies to Enhance Plant Based Functional Foods: An Overview. Front Genet. 2021;12:742095.
- Kato H, Takahashi S, Saito K. Omics and integrated omics for the promotion of food and nutrition science. J Tradit Complement Med. 2011;1(1):25-30.
- McGuire AL, et al. The road ahead in genetics and genomics. Nat Rev Genet. 2020;21(10):581-596.
- Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27(4):626-638.
- Alayande KA, Aiyegoro OA, Nengwekhulu TM, Katata-Seru L, Ateba CN. Integrated genome-based probiotic relevance and safety evaluation of Lactobacillus reuteri PNW1. PLoS One. 2020;15(7):e0235873.
- Dai X, Shen L. Advances and Trends in Omics Technology Development. Front Med (Lausanne). 2022;9:911861.
- Bisanz JE, et al. A Genomic Toolkit for the Mechanistic Dissection of Intractable Human Gut Bacteria. Cell Host Microbe. 2020;27(6):1001-1013.e9
- Pasolli E, De Filippis F, Mauriello IE, Cumbo F, Walsh AM, Leech J, Cotter PD, Segata N, Ercolini D. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat commun. 2020.11(1):2610.
- Wang B, Yao M, Lv L, Ling Z, Li L, The human microbiota in health and disease. Engineering, 2017;3(1):71-82.
- Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. Transcriptomics technologies. PLoS Comput Biol. 2017;13(5):e1005457.
- Wahl D, LaRocca TJ. Transcriptomic Effects of Healthspan-Promoting Dietary Interventions: Current Evidence and Future Directions. Front Nutr. 2021;8:712129.
- Kiu R, et al. Bifidobacterium breve UCC2003 Induces a Distinct Global Transcriptomic Program in Neonatal Murine Intestinal Epithelial Cells. iScience. 2020;23(7):101336.
- Edlund A, Yang Y, Yooseph S, He X, Shi W, McLean JS. Uncovering complex microbiome activities via metatranscriptomics during 24 hours of oral biofilm assembly and maturation. Microbiome. 2018;6(1):217.
- Kwoji ID, Aiyegoro OA, Okpeku M, Adeleke MA. 'Multi-omics' data integration: applications in probiotics studies. NPJ Sci Food. 2023;7(1):25.
- Kussmann M, Panchaud A, Affolter M. Proteomics in nutrition: status quo and outlook for biomarkers and bioactives. J Proteome Res. 2010;9(10):4876-4887.
- Ruiz L, et al. Tackling probiotic and gut microbiota functionality through proteomics. J Proteomics. 2016;147:28-39.
- Bianchi L, et al. A Combined Proteomics, Metabolomics and In Vivo Analysis Approach for the Characterization of Probiotics in Large-Scale Production. Biomolecules. 2020;10(1):157.
- Wu L, et al. Integrated Multi-Omics for Novel Aging Biomarkers and Antiaging Targets. Biomolecules. 2021;12(1):39.
- Ovesná J, et al. High throughput 'omics' approaches to assess the effects of phytochemicals in human health studies. Br J Nutr. 2008;99 E Suppl 1:ES127-34.
- Zhang Z, Wu S, Stenoien DL, Paša-Tolić L. High-throughput proteomics. Annu Rev Anal Chem (Palo Alto Calif). 2014;7:427-54.
- Zhang X, et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nature communications. 2018;9(1):2873.
- Kleikamp HBC, et al. Database-independent de novo metaproteomics of complex microbial communities. Cell Syst. 2021;12(5):375-383.e5.
- Kleiner M, et al. species biomass contributions in microbial communities via metaproteomics. Nat Commun. 2017;16;8(1):1558.
- Pan L, et al. Metabolomic analysis of significant changes in Lactobacillus casei Zhang during culturing to generation 4,000 under conditions of glucose restriction. J. Dairy Sci. 2019;102(5):3851-3867.
- Salminen S, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. 2021;18(9):649-667. doi: 10.1038/s41575-021-00440-6. Epub 2021 May 4. Erratum in: Nat Rev Gastroenterol Hepatol. 2021;Erratum in: Nat Rev Gastroenterol Hepatol. 2022;19(8):551.


